Feb 04, · The obtaining of McFarland suspension is controlled by measurements in a densitometer or spectrophotometer, Reading of Results. the patient with a chemical (such as the antibiotic). The result obtained in such test does not take into account the factors affecting the antibiotic in the patient’s body Trigger factors of development of the CN can also be infections (osteomyelitis precedes CN) and revascularization. A necessary condition for the emergence of CN is the presence of peripheral neuropathy, in particular, diabetic distal sensitive polyneuropathy. Several methods can Sep 03, · These data suggest that EBNA2 can enhance or reduce expression of the gene depending on the risk allele, likely promoting EBV infection. This is consistent with the concept that these MS risk loci affect MS risk through altering the response to EBNA2. Together with the extensive data indicating a pathogenic role for EBV in MS, this study supports targeting EBV and EBNA2 to reduce
The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance
Our website does not fully support your browser. We've detected that you are using an older version of Internet Explorer. Your commerce experience may be limited. Please update your browser to Internet Explorer 11 or above. We use cookies and similar technologies to make our website work, run analytics, improve our website, and show you personalized content and advertising. Some of these cookies are essential for our website to work.
To find out more about cookies and how to manage cookies, read our Cookie Policy. If you are located in the EEA, the United Kingdom, or Switzerland, you can change your factors affecting spectrophotometer reading at any time by clicking Manage Cookie Consent in the footer of our website. Your Account. To protect your privacy, your account will be locked after 6 failed attempts. After that, you will need to contact Customer Service to unlock your account.
You have 4 remaining attempts. You have 3 remaining attempts, factors affecting spectrophotometer reading. You have 2 remaining attempts. You have 1 remaining attempt. Contact Customer Service. Forgot Password? Username not found. This field is required. There was an issue with the password reset process. Please try again or contact Customer Service.
Factors affecting spectrophotometer reading in with Your New Password. You have not verified your email address. A verified email address is required to access the full functionality of your Promega. com account. Resend verification email. Cell Biology. Nucleic Acid Analysis. Human Identification. Molecular Diagnostics. Protein Analysis. Applied Sciences. Drug Discovery. Featured Research Topics. Infectious Diseases.
Custom Manufacturing. Onsite Stocking, factors affecting spectrophotometer reading. Format and QC. Automation Solutions. Custom Factors affecting spectrophotometer reading Development. Student Resources. Peer Reviewed Literature. Product Usage Information. Global Support. Medical Affairs. Local Sales Support. About Promega. Join Our Team, factors affecting spectrophotometer reading. Contact Us. Your Cart. Current Items 0. For ordering information on the products discussed here, please visit our Nucleic Acid Extraction product pages.
Finding a suitable DNA isolation system to satisfy your downstream application needs is vital for the successful completion of experiments. This DNA purification guide addresses general information on the basics of DNA extraction, plasmid preparation and DNA quantitation, as well as how optimized purification techniques can help increase your productivity, so you spend less time purifying DNA and more time developing experiments and analyzing data. There are five basic steps of DNA extraction that are consistent across all the possible DNA purification chemistries: 1 disruption of the cellular structure to create a lysate, 2 separation of the soluble DNA from cell debris and other insoluble material, 3 binding the DNA of interest to a purification matrix, 4 washing proteins and other contaminants away from the matrix and 5 elution of the DNA.
The goal of lysis is to rapidly and completely disrupt cells in a sample to release nucleic acid into the lysate. There are four general techniques for lysing materials: physical methods, enzymatic methods, chemical methods and combinations of the three.
Physical methods typically involve some type of sample grinding or crushing to disrupt the cell walls or tough tissue. A common method of physical disruption is freezing and grinding samples with a mortar and pestle under liquid nitrogen to provide a powdered material that is then exposed to chemical or enzymatic lysis conditions.
Grinders can be simple manual devices or automated, capable of disruption of multiple well plates. Physical methods are often used with more structured input materials, factors affecting spectrophotometer reading, such as tissues or plants.
Other devices use bead beating or shaking in the presence of metallic or ceramic beads to disrupt cells or tissues, or sonication to disrupt tissues and lyse cells.
Chemical methods can be used alone with easy-to-lyse materials, such as tissue culture cells or in combination with other methods. Cellular disruption is accomplished with a variety of agents that disrupt cell membranes and denatures proteins. Chemicals commonly used include detergents e.
Enzymatic methods are often used with more structured starting materials in combination with other methods with tissues, plant materials, bacteria and yeast. The enzymes utilized help to disrupt tissues and tough cell walls. Depending on factors affecting spectrophotometer reading starting material, typical enzymatic treatments can include: lysozyme, zymolase and liticase, proteinase K, collagenase and lipase, among others.
Enzymatic treatments can be amenable to high throughput processing, but may have a higher per sample cost compared to other disruption methods.
In many protocols, factors affecting spectrophotometer reading, a combination of chemical disruption and another is often used since chemical disruption of cells rapidly inactivates proteins, including nucleases. Depending on the starting material, cellular lysates may need to have cellular debris removed prior to nucleic acid purification to reduce the carryover of unwanted materials proteins, lipids and saccharides from cellular structures into the purification reaction, which can clog membranes or interfere with downstream applications.
Usually clearing is accomplished by centrifugation, filtration or bead-based methods. Centrifugation can require more hands-on time, but it is able to address large amounts of debris. Filtering can be a rapid method, but samples with a large amount of debris can clog the filter. Bead-based clearing, like the method used with Promega particle-based plasmid prep kits, can be used in automated protocols, but can be overwhelmed with biomass.
Once a cleared lysate is generated, the DNA can then be purified by many different chemistries, such as silica, ion exchange, cellulose or precipitation-based methods. Regardless of the method used to create a cleared lysate, the DNA of interest can be isolated using a variety of different methods. Promega offers genomic DNA isolation systems based on sample lysis by detergents, and purification by binding to matrices silica, factors affecting spectrophotometer reading, cellulose and ion factors affecting spectrophotometer readingwhich is where interest has primarily been focused in recent years.
Each of these chemistries can influence the efficiency and purity of the isolation, and each have a characteristic binding capacity. Bind capacity is an indication of how much nucleic acid an isolation chemistry can bind before it reaches the capacity of the system and no longer isolates more of that nucleic acid.
We can build design features into these chemistries by manipulating the binding conditions to enrich for different categories of nucleic acid e. This type of chemistry does not rely on a binding matrix, but rather on alcohol precipitation, factors affecting spectrophotometer reading.
Following the creation of lysate, the cell debris and proteins are precipitated using a high-concentration salt solution. The high concentration of salt causes the proteins to fall out of solution, and then centrifugation separates the soluble nucleic acid from the cell debris and precipitated protein 1. The DNA is then precipitated by adding isopropanol to the high-concentration salt solution.
This forces the large genomic DNA molecules out of solution, while the smaller RNA fragments remain soluble. The insoluble DNA is then pelleted and separated from salt, isopropanol and RNA fragments via centrifugation. Additional washing of the pellet with ethanol factors affecting spectrophotometer reading the remaining salt and enhances evaporation.
Lastly, the DNA pellet is resuspended in an aqueous buffer like Tris-EDTA or nuclease-free water and, once dissolved, is ready for use in downstream applications.
The technology for these genomic DNA purification systems is based on binding of the DNA to silica under high-salt conditions 2—4. The key to isolating any nucleic acid with silica is the presence of a chaotropic salt like guanidine hydrochloride.
Chaotropic salts present in high quantities are able to disrupt cells, deactivate nucleases and allow nucleic acid to bind to silica. These washes remove contaminating proteins, lipopolysaccharides and small RNAs to increase purity while keeping the DNA bound to the silica membrane column, factors affecting spectrophotometer reading. Once the washes are finished, the genomic DNA is eluted under low-salt conditions using either nuclease-free water or TE buffer, factors affecting spectrophotometer reading.
Binding to silica is not DNA specific, so if pure DNA is required, factors affecting spectrophotometer reading, there is also the option to add ribonuclease RNase A to the elution buffer. RNA may be may be copurified with gDNA, and the addition of RNase to the elution buffer ensures the removal factors affecting spectrophotometer reading the vast majority of contaminating RNA.
This chemistry can be adapted to either paramagnetic particles PMPslike Promega silica-coated MagneSil® PMPs, or silica membrane column-based formats. While both methods generally represent a good balance of yield and purity, the silica membrane column format is more convenient.
For automated factors affecting spectrophotometer reading, either the well silica membrane plates or the MagneSil® PMPs are easily adapted to a variety of robotic platforms.
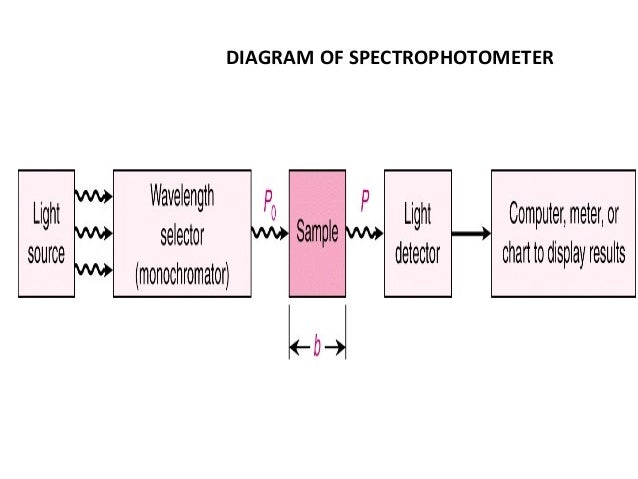
Spectrophotometry (Absorbance)
, time: 6:26Ammonia | US EPA
One of the most critical factors affecting the yield of plasmid from a given system is the copy number of the plasmid. Copy number is determined primarily by the region of DNA surrounding and including the origin of replication in the plasmid. This area, known as the replicon, controls replication of plasmid DNA by bacterial enzyme complexes Factors affecting mathematics teachers’ use of teaching aids in facilitating students learning: The case of Jimma town secondary school mathematics teachers March, Optical density can be used as a measure to indicate the colour of the product sample. A cut-off point above or below a sample is deemed to have passed its shelf life. It should be determined by the specific product requirements. Using an appropriately calibrated spectrophotometer, absorbance of samples are recorded at nm
No comments:
Post a Comment